▎Achievements
Mining Big Data to Reveal the Transcriptional Landscape of Bacteria in Cancer
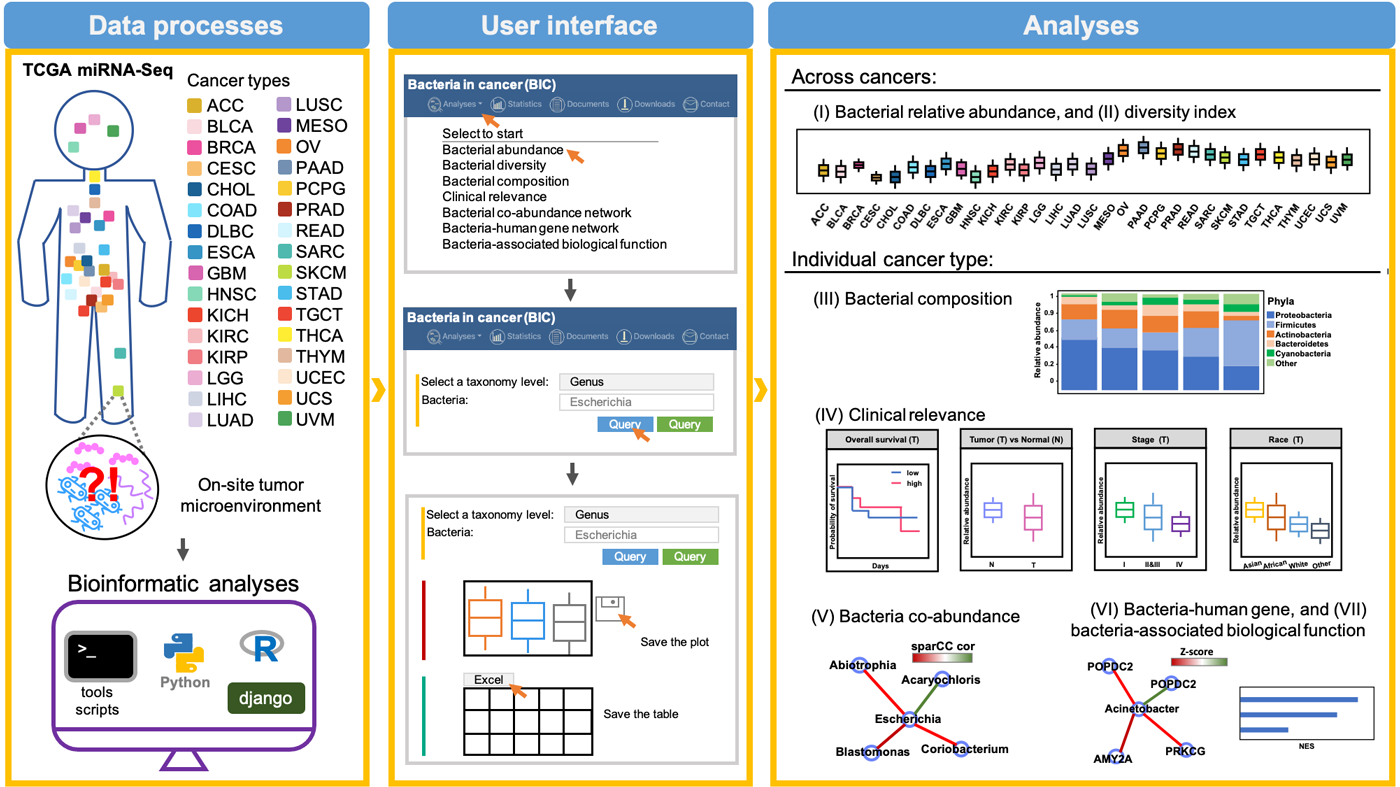
Analyzing the microRNA sequence data of 32 human cancer tissues, mining the implied bacterial transcriptional landscape, and constructing the “BIC” database can provide cancer microenvironment information related to microbial communities.
In the past, internal organs other than the human gut were thought to be sterile. In particular, bacteria detected in tumor tissue were often considered contaminants introduced during sampling. Recently, however, increasing evidence has shown that various microorganisms are present in cancer tissues. Based on the concept of “making the best use of everything,” this study carried out big data analysis of human cancer microRNA sequence data from The Cancer Genome Atlas (TCGA). By reusing the sequences that should have been discarded because they did not correspond to human genes, and performing sequence alignment with bacterial gene sequence data, the information, such as bacterial species and expression quantities present in cancer tissues, were obtained and “BIC” database, which provides biological information about the cancer microenvironment in relation to the microbial community, was then developed. The research results were published in Nucleic Acids Research, the top-ranking journal in biochemical research.
By mining the sequence data from 10,362 patient tissue samples in 32 cancer types and using multiple bioinformatic analyses, the results of cancer-associated bacterial information, including the relative abundance of bacteria, bacterial diversity, associations with clinical relevance, the co-expression network of bacteria and human genes, and their associated biological functions, were acquired. BIC database provides an online interface for query and visualization so that users can quickly and effectively use and download this information. BIC is a public database that users can access freely, and all the developed source codes are available on GitHub. Researchers and enthusiasts in the bioinformatics-related community are also welcome to develop other applications accordingly.
The first author, Kai-Pu Chen, is a Ph.D. student of the Graduate Institute of Biomedical Electronics and Bioinformatics. Chen is diagnosed with the rare disease Spinal Muscular Atrophy (SMA). The research was completed thanks to NTU and the Department of Life Sciences for providing accessible spaces on campus for students such as Chen, so they can do their best work without worry.
This study was jointly conducted by Dr. Hsueh-Fen Juan, Distinguished Professor of the Department of Life Sciences and Graduate Institute of Biomedical Electronics and Bioinformatics and Director of the Center for Computational and Systems Biology, and Dr. Hsuan-Cheng Huang, Professor of the Institute of Biomedical Informatics at National Yang Ming Chiao Tung University. This work was supported by the Ministry of Science and Technology, Taiwan, the Ministry of Education, and the National Center for High-performance Computing (NCHC), which provided computational and storage resources. The research members include Dr. Chia-Lang Hsu, Associate Research Fellow of the Department of Medical Research of National Taiwan University Hospital, and Dr. Yen-Jen Oyang, Professor of the Graduate Institute of Biomedical Electronics and Bioinformatics.


